Given two curves $y1(x)$ and $y2(x)$ how can we find the best overlap of those two curves ?
The notion of "best overlap" refers to the minimisation of a distance between the curves. Definin the distance is thus the first step.
In the following examples, we will take the distance between two curves as the quadratic difference.
Provided that $y1$ and $y2$ have the same number of points $N$, we define the distance as follows:
$$d(y1, y2) = \sum_{i=0}^{N-1} \left[y_1(x_i) - y_2(x_i) \right]^2 $$Now, we supposed that the the curves are considered at the same locations $x_i$, so we will just use the index $i$ to tag the values of y in a array of length $N$:
$$d(y1, y2) = \sum_{i=0}^{N-1} \left(y_{1,i} - y_{2,i} \right)^2 $$Intuitively, finding the best overlap would just consist in making a curve "glide" horizontally while keeping the other fixed. Then, for each increment of translation, we would compute the distance and try to get the minimum among all the computed distances. This would give us the "best" overlap.
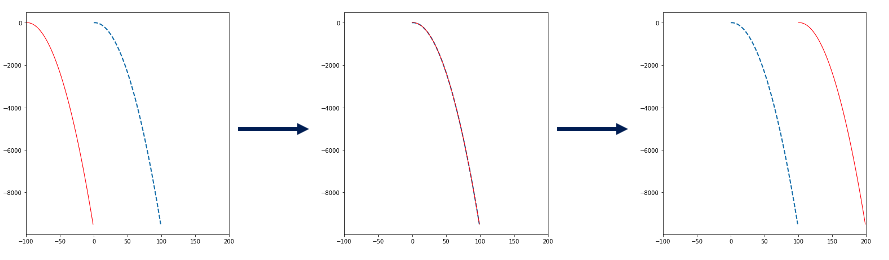
Here I would like to mention two things:
We may be tempted to go for a convolution product because it seems to be exactly what we are trying to do. However, as I will show later it doesn't work.
The problem that will come quickly is how to compare the distance between two sets of curves, where both sets are of different lengths (number of points). We need to renormalize it, otherwise, a bad overlap between two short pieces may yield a better distance than a good yet non perfect overlap between two long pieces of curve.
We will call $d_{m}$ the normalized distance and define it as follows:
$$d_{m}(y1, y2) = \frac{d(y1,y2)}{N} = \frac{1}{N} \sum_{i=0}^{N-1} \left(y_{1,i} - y_{2,i} \right)^2 $$